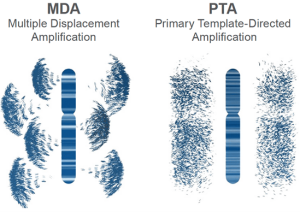
Single-cell genomics has transformed our understanding of cellular diversity in both health and disease. Yet, a key technical challenge persists: how to amplify the minute amounts of DNA in a single cell without introducing errors or bias. For years, researchers have relied on Multi-Displacement Amplification (MDA) as the gold standard. But a newer method, Primary Template-directed Amplification (PTA), developed by BioSkryb, promises to dramatically improve the quality of single-cell genomic data. In this post, we’ll explore how PTA compares to traditional MDA and why it might be the new go-to technique for researchers pushing the limits of genomic resolution.
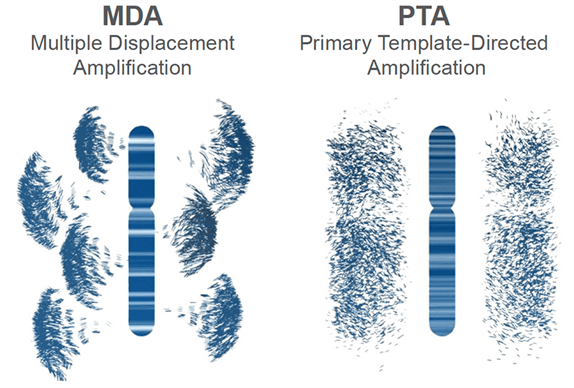
What is multi-displacement amplification (MDA)?
MDA is an isothermal amplification method that uses the high-fidelity phi29 DNA polymerase to amplify genomic DNA. It’s widely used for single-cell applications due to its strong strand-displacement activity. MDA can generate micrograms of DNA from just picogram inputs.
Strengths of MDA:
- High amplification yield
- High-fidelity polymerase
- Isothermal (no need for thermocycling)
Limitations of MDA:
- Amplification bias: some genomic regions are preferentially amplified, leading to uneven coverage.
- Allelic dropout: one allele of a heterozygous site may be missed entirely.
- Chimeric artifacts: non-contiguous regions may be mistakenly joined during amplification.
- Poor reproducibility: inconsistent results across cells and experiments.
How PTA works: BioSkryb’s breakthrough
BioSkryb’s Primary Template-directed Amplification (PTA) was designed to overcome the pitfalls of MDA. PTA also uses phi29 polymerase but incorporates a proprietary priming and amplification strategy that emphasizes amplification directly from the primary DNA template, rather than from its amplified products. . It does this by introducing a termination base to stop replication of the same gene over and over again, and thus offers advantages in terms of accuracy, uniformity, and reduced bias compared to MDA.
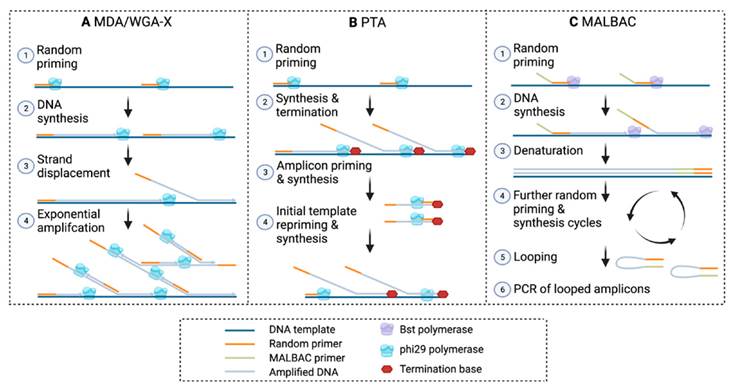
Key Innovations in PTA:
- Linear amplification bias: maintains fidelity to the original genome structure.
- Reduced noise: fewer chimeric and nonspecific products.
- Improved variant calling: enables confident detection of SNVs and CNVs at single-cell resolution.
- Greater uniformity: more even genome coverage, with reduced allelic dropout.
PTA vs MDA
| Feature | MDA | PTA |
|---|---|---|
| Coverage uniformity | Low | High |
| Allelic dropout | High | Low |
| SNV calling accuracy | Moderate | High |
| CNV resolution | Limited | Robust |
| Chimeric artefacts | Common | Rare |
| Ease of use | Established protocols | BioSkryb-specific kits |
| Applications | Single-cell WGS/WGA | Single-cell WGS/WGA, CNV/SNV analysis & whole transcriptome sequencing |
Real-world impact
PTA has already been shown to enable high-confidence single-nucleotide variant (SNV) detection and structural variant mapping in individual cells - applications where MDA often falls short. For instance, in cancer research, where understanding clonal evolution at the single-cell level is essential, PTA allows researchers to detect low-frequency variants and subtle genomic rearrangements that MDA might miss or distort.
The future of PTA in single-cell genomics
While MDA has served the research community well, the limitations of its amplification fidelity, especially for high-resolution single-cell genomics, are becoming increasingly evident. BioSkryb’s PTA method represents a significant step forward, enabling more accurate, reproducible, and comprehensive insights from single cells.
For researchers working at the cutting edge of genomics - whether in oncology, neurology, immunology, or developmental biology - PTA is quickly becoming the new standard.
Curious how PTA could benefit your single-cell project? Contact us to learn more or explore BioSkryb’s range for high-fidelity whole-genome amplification.
